Studies on Stem Cells Research and Therapy
Cell-Penetrating Peptides as a Tool to Deliver Biologically Active Recombinant Proteins to Generate Transgene-Free Induced Pluripotent Stem Cells
1Laboratory for Stem Cell Engineering and Regenerative Medicine (SCERM), Department of Biosciences and Bioengineering, Indian Institute of Technology Guwahati (IITG), Guwahati 781039, Assam, India
2National Centre for Cell Science (NCCS), NCCS Complex, Savitribai Phule Pune University Campus, Ganeshkhind, Pune 411007, Maharashtra, India
3†These authors contributed equally to this work
Author and article information
Cite this as
Dey C, Narayan G, Krishna Kumar H, Borgohain MP, Lenka N, et al. (2017) Cell-Penetrating Peptides as a Tool to Deliver Biologically Active Recombinant Proteins to Generate Transgene-Free Induced Pluripotent Stem Cells. Stud Stem Cells Res Ther. 2017; 3(1): 006-015. Available from: 10.17352/sscrt.000011
Copyright License
© 2017 Dey C, et, al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Delivery of biologically active recombinant proteins using cell-penetrating peptides is a useful tool for transduction of molecular cargo into cells. This protein transduction technology is useful to understand the biological function of a specific protein of interest. Hence, it has been recently employed by severalgroups to understand the molecular functions of stem cell-specific transcription factors inpluripotent stem cells. Pluripotent stem cells have an indefinite self-renewal capacity and can be directed to differentiate into any desired cell type of an adult organism. The groundbreaking discovery of induced pluripotent stem (iPS) cells has revolutionized the field of stem cell research due to its immense potential in in vitro disease modeling, drug screening and regenerative medicine. Although, virus-based gene delivery approaches commonly used to generate iPS cells are robust and highly efficient, however, these methods involve permanent genetic modifications due to viral integrations leading to malignant transformation. Therefore, a protein-based approach to deliver biologically active recombinant proteins in somatic cells to generate iPS cells is safe and will improve the prospects of these cells from bench-to-bedside. This review provides an overview of protein-based somatic cell reprogramming to generate transgene-free iPS cells and gives a glimpse of the bottlenecks associated with this technology.
CPP, cell-penetrating peptide; ES, embryonic stem; FCS, fetal calf serum; GFP, green fluorescent protein; HDAC, histone deacetylase; iPS, induced pluripotent stem; MEF, mouse embryonic fibroblast; NLS, nuclear localization signal/sequence; OSKM, Oct4, Sox2, Klf4, c-Myc; p-iPS, protein-induced pluripotent stem; RFs, reprogramming factors; RFP, red fluorescent protein; SR, serum replacement; TAT, trans-activator of transcription; TEV, Tobacco etch virus; TLR3, Toll-like receptor 3; VPA, valproic acid
Introduction
Pluripotent stem cells such as embryonic stem (ES) and induced pluripotent stem (iPS) cells are elegant in vitro models for exploring cell fate, commensurate with the developmental hierarchy operational during early embryogenesis. Undoubtedly, their enormous potential in regenerative medicine has revolutionized the field of biology and medicine alike. iPS cells which closely resemble ES cells, are unspecialized cells that have two unique characteristics: self-renewal, the ability to proliferate indefinitely; and pluripotency, the ability to differentiate into numerous specialized cell types of all the three germ layers. These unique characteristics of iPS cells make them promising for autologous cell transplantation, that are devoid of immunological concerns and ethical problems unlike that in case of human ES cells. In addition, these cells can also be used for modeling a disease carrying genetic mutation(s) in a cell culture dish as well as for drug toxicity screening applications.
The remarkable discovery of derivation of iPS cells from adult somatic cells by ectopic expression of stem cell-specific transcription factors [1] has opened new avenues for therapeutic applications of stem cells. This depends on the availability of clinical grade human iPS cells that are not constrained by ethical, technical or immunological considerations. Several integrating (e.g. retro- and lenti-viruses) and non-integrating (adeno- and sendai-viruses, piggybac transposons, episomal and polycistronic plasmid vectors) approaches have been successfully used to generate iPS cells [2,3]. Integrating approaches are efficient and robust in reprogramming somatic cells to iPS cells than non-integrating approaches; however, the clinical applicability of such iPS cells is restricted due to the integrated transgenes carrying the risk of insertional mutagenesis and tumor formation [4]. Although, non-integrating approaches reduce genome integration, the DNA of reprogramming cocktail of transcription factors and short vectors after removal can also result in insertional mutagenesis. To circumvent these problems, a virus- and DNA-free safe non-integrative approach of reprogramming is highly desirable. Till date, the promising approaches to generate clinical grade iPS cells are synthetic mRNAs [5,6], small molecules [7], microRNAs [8,9] and recombinant proteins [10-14], which eliminate the possibility of genome alteration by introduced foreign DNA. One of the most fascinating approaches is the transduction of biologically active cell-permeable recombinant proteins, as it provides precise control over dosage, time of application, and gives flexibility to toy with sequence of transcription factor stimulation.
Protein transduction via cell-penetrating peptides (CPPs) is an approach to deliver recombinant proteins and various small and large bioactive molecules into cells, and for this reason, this method has the possibility to replace virus-based gene delivery approaches. In this review, we summarize recent published reports on protein-based somatic cell reprogramming to generate iPS cells and highlight the bottlenecks of this technology.
Cell-penetrating peptides
Cell-penetrating peptides (CPPs), also known as protein transduction domains (PTDs) facilitate the delivery of recombinant proteins, peptides, oligonucleotides and other vital molecules into different mammalian cells. Most of the CPPs have been reported to be innocuous and do not impede the functionality of the biomolecules delivered across the cell membrane. Therefore, these CPPs have a wide application in many research areas, including cancer, apoptosis, cell cycle, neurobiology, signal transduction, and many others [15].
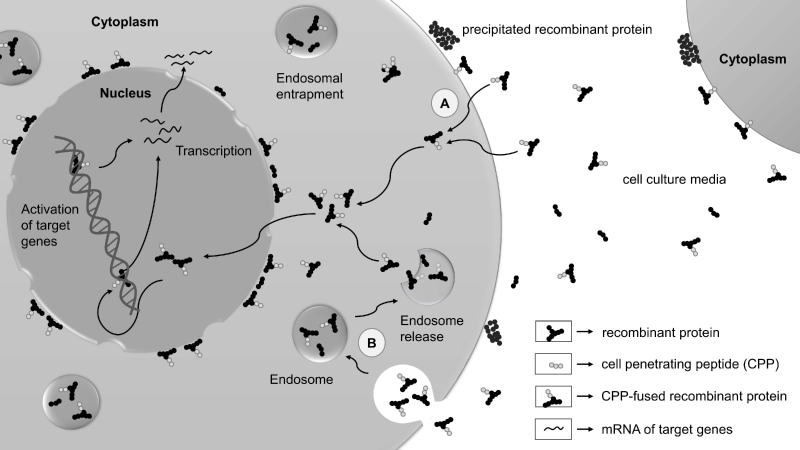
CPPs are a valuable class of short peptides with 3-30 positively charged (basic) amino acids, having an ability to translocate across the cell membranes with limited toxicity. Biologically active recombinant proteins when fused in frame with CPPs are translocated into cells via two different entry mechanisms: energy-independent direct penetration and energy-dependent endocytosis (Figure 1) [16]. However, the detailed CPP uptake mechanisms are largely unclear. This may vary substantially according to several parameters such as CPP, CPP length, CPP-cargo, cell types, cell density, concentration, incubation time and temperature, physicochemical parameters and other reasons [15]. Identifying and optimizing the right parameters will overcome the obstacles that might help in the easy uptake of these CPPs by the cell.
Cell penetrating capability of the trans-activator of transcription (TAT) protein of the Human Immunodeficiency Virus (HIV) was originally identified by two independent groups [17,18]. Subsequent studies demonstrated a short sequence in the basic domain of TAT protein (47-57), now recognized as TAT, can rapidly translocate into the cells more efficiently than full length protein and accumulate in the nucleus [19-21]. Later, Penetratin, a CPP comprising of 16 amino acids, was identified in the third helix of the homeodomain of transcription factor Antennapedia of the fruitfly Drosophila melanogaster [22-24]. Both TAT and Penetratin are rich in arginine and lysine residues imparting a high net positive charge at physiological pH. Recombinant proteins fused to these CPPs or synthetic CPPs rich in arginines can easily get internalized into cells. Generally, 6 to 12 poly-arginines exhibit transduction activity as CPPs [25,26], but as recently reported, even three oligo-arginines (3R) are sufficient to deliver transcription factors into the cells [27]. Since the first discovery more than 25 years ago, several short polycationic (Table 1) and amphipathic CPPs [28] have been identified subsequently. Importantly, even after cellular uptake these recombinant proteins are biologically active. This opens up new possibilities for intracellular delivery of various biologically active molecules into mammalian cells. The ability of these molecular delivery vehicles to translocate different small and large bioactive molecules into cells makes them promising candidates for transport of recombinant proteins, peptides, nucleic acids, liposomes, nanoparticles, medical imaging agents and drug delivery applications.
CPP-mediated protein transduction to generate iPS cells
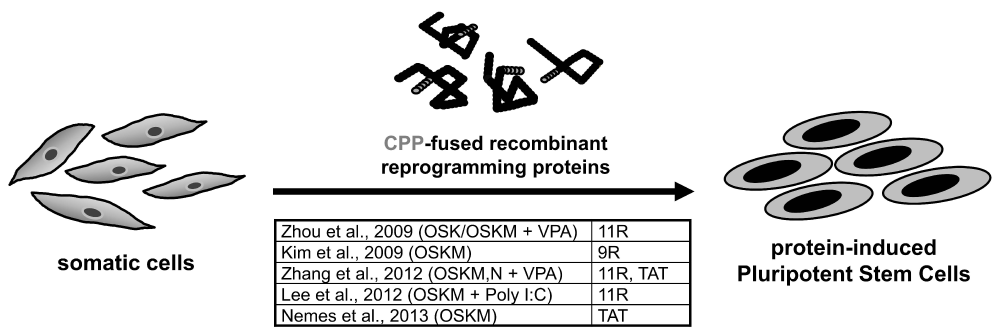
Numerous integrating and non-integrating approaches have been employed to successfully derive iPS cells. Non-integrating approaches entirely replace gene delivery during the reprogramming process. These strategies are of major importance as they do not alter the host genome and generate clinically safe iPS cells. One of the most attractive approaches is the direct delivery of CPP-fused cell permeable recombinant proteins. This strategy has been employed by various studies to understand the role of growth factors and stem cell-specific transcription factors in recombinant forms to understand the biological function in pluripotent stem [29-34] and somatic [35] cells. In addition, this strategy has been recently used by few groups to induce ES cell gene expression program by delivering recombinant reprogramming proteins into the somatic cells to generate transgene-free iPS cells (Figure 2). This approach offers precise control over the dose and time of application, and gives complete flexibility to try and test different combinations of reprogramming factors to unravel their role at different stages during the reprogramming process. These CPP-mediated recombinant reprogramming proteins were delivered at a specific concentration, transduction cycle and exposure time on either mouse or human somatic cells in different studies to generate protein-iPS (p-iPS) cells (Table 2).
The very first study to report the generation of p-iPS cells was by reprogramming mouse embryonic fibroblasts (MEF) using a cocktail of recombinant cell-permeant reprogramming factors, Oct4, Sox2, Klf4 and c-Myc (hereafter, OSKM) [11], otherwise also known as “Yamanaka factors” originally identified by Takahashi and Yamanaka [1]. These reprogramming factors were tagged to CPP, poly-arginines (11R), at the C-terminal end and expressed in E.coli [11]. Recombinant proteins (Oct4-11R, Sox2-11R, Klf4-11R and cMyc-11R) at concentration 0.5-8 µg/ml were observed to translocate into the nucleus and were stable upto 48 hours [11]. For this reason, OG2/Oct4-GFP (green fluorescent protein) reporter MEF cells were treated in four transduction cycles at every 48 hours at concentration 8 µg/ml for 12 hours [11]. However, this did not yield any iPS cell colonies [11]. The reason could be that large amount of these recombinant proteins got trapped inside the endosomes. Hence, only a small fraction of the recombinant reprogramming proteins could translocate into the nucleus. Concurrently, 1 mM Valproic Acid (VPA), a histone deacetylase (HDAC) inhibitor, was also added to the reprogramming proteins cocktail [11], which was earlier reported to significantly improve reprogramming efficiency [36,37]. Application of VPA prevents histone deacetylation by inhibiting HDACs, inducing an open chromatin structure that permits reprogramming factors to generate an ES cell-specific gene expression profile. Inclusion of VPA in the four reprogramming cocktail yielded only 3 GFP +ve iPS cell colonies, whereas only one GFP +ve colony in case of three reprogramming cocktail (Oct4-11R, Sox2-11R and Klf4-11R) at day 30-35 [11]. Like mouse ES cells, these p-iPS cells expressed all pluripotency markers and successfully tested for the pluripotency assays [11]. However, the reprogramming efficiency in this study was very low compared to viral approaches, primarily due to inadequate translocation of reprogramming factors to the nucleus, whereas the majority got confined to endosomes as observed in immunostaining images. Agents stimulating endosomal release of reprogramming proteins will help to improve reprogramming efficiency. Moreover, we hypothesize the masking of nuclear localization signal/sequence (NLS) in recombinant reprogramming proteins, and hence an additional NLS fusion would help them to easily translocate to the nucleus.
Simultaneously, another study reported successful generation of p-iPS cells by reprogramming human fibroblasts using OSKM fused to poly-arginines (9R) [10]. Preliminary experimental designs were based on examining whether the red fluorescent protein (RFP) when fused to 9 poly-arginines (RFP-9R) was able to penetrate into COS-7 cells and human newborn fibroblasts (HNF). Encouraging results were observed showing effective delivery of RFP-9R into both the cell types within few hours, even with the treatment of whole cell extracts [10]. Based on this data, stable HEK293 cell lines were generated expressing each of the four reprogramming factors fused with 9R [10]. These stable cell lines showed high expression of these proteins as determined by western blot analysis. Further treatment of HNF with the cell extracts from HEK293 resulted in efficient intracellular translocation of the recombinant proteins within 8 hours [10]. However, single dose of protein transduction did not generate any iPS cell colony, therefore, repeated treatments for six consecutive days for 16 hours was performed [10]. This eventually gave rise to p-iPS cell colonies after 8 weeks, but with low reprogramming efficiency (0.001%) and slow kinetics (8 weeks) [10]. These p-iPS cell colonies generated in this study were indistinguishable from human ES cells in morphology, proliferation and expression of pluripotency markers [10]. The low efficiency and kinetics could be due to treatment with whole cell extracts from the expressing cells thereby limiting the concentration of reprogramming factors delivered into target cells. Hence, treatment of fibroblasts with purified reprogramming factors might facilitate generation of iPS cells with higher efficiency and kinetics.
Reprogramming of mouse or human somatic cells to generate iPS cells using CPP-mediated recombinant protein transduction approach has remained highly inefficient [10,11]. Addressing this problem, a fascinating study recently reported that Toll-like receptor 3 (TLR3) signaling enables efficient induction of pluripotency due to the innate immune response to viral or modified mRNA [12]. However, this vital signaling is not activated in CPP-mediated protein transduction, which could be the probable reason for its low reprogramming efficiency. To activate this signaling, addition of TLR3 agonist (Polyinosinic-polycytidylic acid (Poly I:C)) to CPP-fused reprogramming cocktail (OCT4-11R, SOX2-11R, KLF4-11R, and c-MYC-11R) resulted in histone acetylation, down-regulating expression of various HDACs (HDAC 1,2,5,7) in CPP-treated human fibroblasts to generate p-iPS cells with high reprogramming efficiency and faster kinetics [12]. Therefore, TLR3 stimulation by Poly I:C is critical for activation of downstream target genes of the reprogramming factors and this TLR3 activation may trigger rapid and global changes in the expression of epigenetic modifiers that promote an open chromatin configuration and favor nuclear reprogramming [12]. Screening highly efficient novel TLR3 agonists that activate innate immune response would be desirable to improve efficiency and kinetics of CPP-mediated protein-based somatic cell reprogramming and further enhance the robustness of this technology.
Using macromolecule intracellular transduction technology (MITT), Lim et al. applied to deliver biologically active reprograming factors OSKM and NANOG (Group A) or OSKM and LIN28 (Group B) into human skin fibroblasts to generate iPS cells [38]. This technology is based on the ability of hydrophobic macromolecule transduction domains (MTDs) to facilitate bidirectional transfer of recombinant proteins and peptides through the cell membrane. Seven MTDs were selected on the basis of high transduction ability. They were tagged to the reprogramming factors using a 6-histidine tag and nuclear localization signal/sequence (NLS) for nuclear localization [38]. However, neither Group A nor Group B gave rise to stably expandable iPS cell colonies, even though the colonies resembled morphologically to ES cells [38]. Hence, further investigation and optimization of this technology is required to deliver reprogramming factors to generate stably expandable human p-iPS cells.
Comparing p-iPS cell generation from two different genetic backgrounds, Nemes et al. attempted to generate mouse iPS cells using recombinant proteins from inbred (C57BL6) and outbred (ICR) strains [14]. They linked TAT with recombinant proteins to facilitate cell penetration. In addition, they fused NLS to promote nuclear delivery, thereby minimizing the quantity of proteins getting trapped in endosomes, and eventually preventing their degradation by hydrolytic enzymes. A short linker between TAT and NLS was also added to provide the necessary flexibility to the protein structure. MEFs from inbred and outbred strains were treated with recombinant TAT-fused OSKM for four times with an interval of 48 hours [14]. Nevertheless, only the outbred MEFs were successfully reprogrammed [14]. The possible reason, in addition to lower efficiency of protein transduction, could be the variation in the effect of modifier genes from two different genetic backgrounds [39]. Generated p-iPS cells of outbred strain were positive for pluripotency markers and qRT-PCR analysis expressed a small increase in the level of nuclear and membrane markers in the cell line [14]. Further to investigate iPS cell generation at different O2 levels, they performed cell reprogramming at atmospheric and low (5%) O2 levels [14]. iPS cell colonies were obtained in both genetic backgrounds, however, these colonies were not stable as they differentiated after two to three passages [14]. This could be mainly due to low stability of recombinant reprogramming proteins and/or poor endosomal release of these proteins after cellular uptake, which ultimately leads to incomplete reprogramming.
To overcome these major bottlenecks, Thier et al., showed how media conditions could be optimized to achieve enhanced stability and endosomal release to maximize entry of biologically active protein into somatic cells [40,41]. In two separate studies, they have purified recombinant Oct4 and Sox2 fused to NLS, TAT and Histidine (His) tag at the C-terminus from E.coli, namely Oct4-TAT [40] and Sox2-TAT [41] respectively. Purified Oct4-TAT and Sox2-TAT showed limited solubility under cell culture conditions and their stability was found to be compromised [40,41]. Previously serum was used to increase the stability of the recombinant fusion proteins [42,43]. To improve Oct4-TAT and Sox2-TAT stabilization, serum or serum replacement (SR) supplemented with lipid-rich albumin fraction (Albumax) was tested [40,41]. Though SR increased the transduction efficiency than serum, but it compromised with the stability of the proteins. Moreover, the long-term activity of Oct4-TAT and Sox2-TAT was much improved when fetal calf serum (FCS), SR and Albumax were used together at a specific proportion. After detailed examination, 2.5% of FCS and Albumax (2.5%) for stability, and 7.5% of SR for transduction efficiency was found to be optimal [40,41]. This combination unblocks transduction inhibiting effects of FCS and provides better transduction efficiency and stability. Ultimately these supplements satisfy the criteria needed for the successful protein transduction media conditions for reprogramming. Even though the stability of recombinant proteins was improved, the endosomal entrapment of these proteins became another huge barrier to its successful delivery. Lysomotrophic agents that affects the endosomal integrity such as sucrose, was used to enhance the endosomal release of Oct4-TAT [40]. Apart from the enhancement of endosomal release, sucrose also facilitates the concentration of purified Oct4-TAT up to 500 µg/ml. Using “3 viruses + 1 protein” assay, where Oct4 virus was substituted by Oct4-TAT recombinant protein and applied to SKM infected MEF cells under optimal media conditions, it showed reprogramming efficiency in the same order of magnitude to OSKM viral transduction [40]. However, the reprogramming efficiency was one order lower in case of Sox2-TAT replacing Sox2 virus in reprogramming OKM infected MEF cells [41]. This could be because no sucrose was used to facilitate release of Sox2-TAT recombinant protein from the endosomes. In addition, the efficacy of reprogramming directly depends on the concentration and exposure time period of Oct4-TAT and Sox2-TAT. Both Oct4-TAT- and Sox2-TAT-iPS cells expressed pluripotency markers and differentiated into all three germ layers [40,41]. Further animal studies are needed to be carried out for final confirmation of pluripotency in vivo.
Employing the same “3 viruses + 1 protein” assay, recombinant Klf4 coupled with CPP was used to generate iPS cells from OSM infected MEF cells [44]. TAT and Penetratin were used to deliver recombinant Klf4 in this study and various combinations of CPPs, Klf4 and a reporter Discosoma red fluorescent protein (DsRed) (TRK, TR, TK, KT, PRK, PR and PK constructs) were evaluated for maximum reprogramming efficiencies. The TAT peptide fused with C-terminus of Klf4 (KT) showed reprogramming activity, unlike that fused with N-terminus (TK) [44]. Similarly, low reprogramming efficiency was observed, due to cationic property of Penetratin in which it was fused with N-terminus of Klf4 (PK). This could be one reason why Pan et al. also failed to generate human iPS cells due to N-terminal direct fusion of TAT to reprogramming factors OSKM [45]. Direct linking of TAT/Penetratin to N-terminus negatively interferes with the clusters of acidic residues rich region of Klf4 which interacts with p300/CBP coactivators, components of the histone acetyltransferase complex [46]. Presumably, this results in masking of transcription activation domain of Klf4 due to the strong cationic/amphipathic property of TAT/Penetratin leading to poor reprogramming activity. Generation of the constructs with DsRed between TAT and Klf4 (TRK) exhibited maximum reprogramming efficiency as it unveiled the transcriptional activation domain of Klf4 [44]. However, this enhanced reprogramming efficiency was only due to a small amount of recombinant TRK entering the nucleus, as most of the TRK transduced was localized in cytoplasm and/or peri-nuclear areas. Initially it was assumed that cytoplasmic localization was due to inappropriate construct design. Later it was found that inadequate intracellular trafficking due to misfolding of recombinant Klf4 and limited endosomal release could be the cause. Hence, it represents one of the major bottlenecks for the recombinant protein transduction as reported and observed in immunostained images of different studies [35,45,47-52]. Overall, Klf4 transduction mediated reprogramming directly depends on dose, frequency, transduction cycles and exposure time. Here they reported that four cycles with 72 hours transduction interval of 50nM TRK proteins were required for an effective generation of iPS cells from somatic cells [44]. Essentially, Klf4 concentration used here was 3-fold lower than that used by Zhou et al., 2009. In addition, reprogramming time to generate iPS cells was less with Klf4 protein transduction than that required by Klf4 substituted small molecule, Kenpaullone [44]. Importantly, TRK proteins can even further enhance the reprogramming efficiency if the major bottlenecks such as cytoplasmic/peri-nuclear localization and endosomal entrapment are thoroughly addressed.
Poly-arginines (9R/11R) and TAT have been commonly used by different studies for p-iPS cell generation [10-12,14,40,41,44]. To compare these two commonly used CPPs for cell reprogramming, Zhang et al. carried out protein-based reprogramming of somatic cells and compared the efficiency of TAT fused reprogramming factors (TAT-RFs) with poly-arginines (11R) fused reprogramming factors (11R-RFs) [13]. Incubation time and concentration for efficient transduction was tested and was found to be 2 hours and 25-400 nM respectively for three TAT-RFs, TAT-Klf4, TAT-Sox2 and TAT-c-Myc. Treatment of TAT-Oct4 at higher concentrations caused reduced transduction and cell death, and therefore 5-50 nM was found to be the suboptimal concentration. Notably, they demonstrated that TAT-RFs were transcriptionally more active than the respective 11R-RFs in activating their target genes. This could be mainly due to significant positive charges in 11R CPP which may negatively interfere with transcriptional activation domains of eukaryotic transcription factors often rich in acidic amino acids. This indicated that TAT CPP could be a better choice than 11R for protein-based iPS cell generation [13]. Further, the reprogramming activity of TAT-RFs were investigated using “3 viruses + 1 protein” assay. The addition of TAT-RFs increased the efficiency to multiple folds as compared to the untreated cells. This indicated that TAT-RFs can replace, but, were found to be less transcriptionally active when compared to their respective retrovirally transduced RFs [13]. For this reason, these TAT-RFs failed to reprogram human foreskin fibroblasts (HFF) to generate human iPS cell colonies [13]. Further improvements can be made in the design of their constructs by adding a linker or a reporter gene between TAT and reprogramming factor(s) providing flexibility. Tagging TAT at the C-terminal end would also help to improve the reprogramming efficiency with mouse fibroblasts and also to generate human p-iPS cells. Direct tagging of TAT to reprogramming factors at the N-terminal end has been observed to cause hindrance as reported earlier [14,40,44,41,52]. NLS fusion would also help to promote nuclear localization, reducing protein degradation by the acidic pH and digestive hydrolytic enzymes in endosomes. Human iPS cell colonies were obtained only after inclusion of TAT-Nanog and 125 μM VPA in the four reprogramming cocktail, having efficiency and kinetics at par with four retroviruses alone [13].
Strategies to improve protein transduction and biological activity
The ultimate aim of CPP-mediated protein transduction is to deliver desired biological activity of transduced proteins without affecting the viability of the transduced cells. To improve biological activity, Konno et al. demonstrated that removal of CPP from intracellularly delivered recombinant reprogramming proteins can help regain lost or reduced biological activity [53]. Presence of CPPs in reprogramming proteins is known to interfere with proper folding inside the cells and thereby decreasing the biological activity. In order to overcome this problem, they employed a site-specific protease derived from tobacco etch virus (TEV), to excise out the CPPs after the intracellular delivery of reprogramming proteins [53]. For this, the authors fused poly-arginines (11R) with reprogramming proteins, Sox2 and Oct4. TEV protease recognition site was inserted between 11R and Sox2 or Oct4 [53]. Further, they showed that deletion of CPPs from reprogramming proteins by TEV protease after delivery into the cells substantially restored their biological activity, whereas the presence of CPPs compromised their functions [53]. Moreover, they discovered that TEV proteases did not affect the endogenous proteins; therefore, ES cell identity remained unaltered [53]. This study clearly demonstrated that CPPs interfered with the biological activity of reprogramming proteins, which can be restored by intracellular excision of CPP using a specific protease. The application of this strategy in derivation of p-iPS cells is anticipated to improve both reprogramming efficiency and kinetics, and work efficiently with cell lines which are challenging to reprogram.
Recently, Ryu and colleagues, employed a novel CPP of non-viral origin to increase efficiency of intracellular delivery of reprogramming factors by overcoming the issues related to their solubility and stability [52]. They engineered Bombyx mori 30Kc19 CPP from silk and tagged it to the C-terminus of the reprogramming proteins, generating Oct4-30Kc19, Sox2-30Kc19, Klf4-30Kc19, c-Myc-30Kc19 and L-Myc-30Kc19 constructs [52]. They discovered that 30Kc19 CPP conjugated reprogramming factors exhibited improved soluble expression and stability in vitro. In contrast, unconjugated reprogramming factors aggregated in the inclusion bodies [52]. The authors concluded that enhanced solubility of recombinant reprogramming proteins was due to the C-terminus fusion of the CPP [52]. Similarly, increased stability of reprogramming proteins was attributed to the shielding effect of 30Kc19 [52], without addition of lipid-rich albumin supplements or serum to the cell culture media. Further, on incubation with human dermal fibroblasts (HDFs), they found that the rate of intracellular delivery and transcriptional activity of these reprogramming proteins were remarkably enhanced [52]. However, immunofluorescence images showed that still a large amount of proteins were trapped in endosomes and also observed in the peri-nuclear region [52]. These proteins can be released from endosomes using lysomotrophic agents such as chloroquine, ammonium chloride, sucrose, or pH-sensitive fusogenic hemagglutinin-2 (HA2) subunit peptide [54]. Treatment with these agents will result in the endosomal release of these reprogramming proteins increasing the amount of these proteins entering the nucleus, further improving the transcriptional activity. Many groups have also demonstrated delivery of CPP-cargo into the nucleus with an additional NLS to overcome peri-nuclear entrapment [14,38,40,41,50]. Therefore, 30Kc19 can be used as a potential non-toxic carrier of non-viral origin enhancing solubility, stability and transcriptional activity of recombinant reprogramming proteins. Further role in generation of p-iPS cells by delivering these recombinant reprogramming proteins in active form fused to this CPP is yet to be investigated.
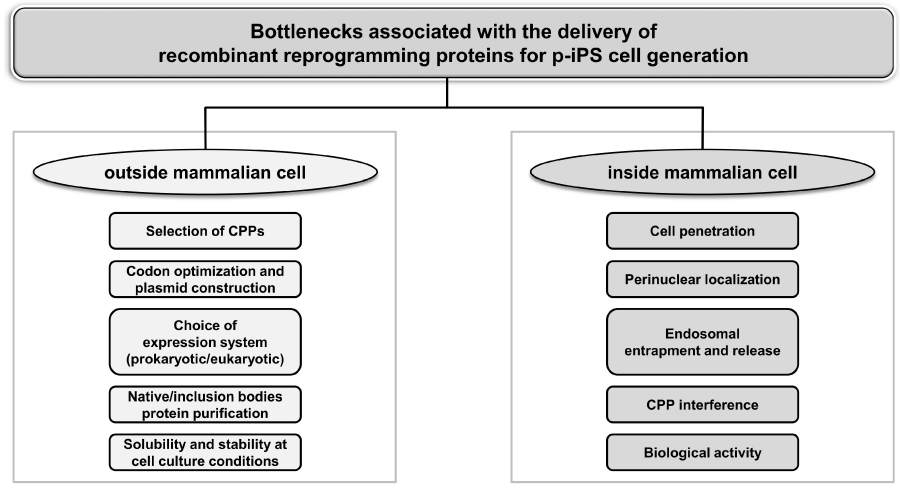
As discussed earlier and highlighted in Figure 3, several bottlenecks associated with CPP-mediated protein transduction to deliver reprogramming factors are required to overcome for successful generation of p-iPS cells. The studies listed above addressed only few of these major bottlenecks in their experimental design. As a result, these studies failed to generate stable p-iPS cells using only OSKM reprogramming factors [11,14,38,45] or resulted in poor reprogramming efficiency [10]. Subsequently, inclusion of mutagenic small molecules such as VPA [11,13] and/or additional reprogramming factors such as Nanog [13] has been used to generate p-iPS cells. A comprehensive study addressing all the bottlenecks is required to successfully generate p-iPS cells with high reprogramming efficiency and faster kinetics, at par with robust and highly efficient viral approaches. Combinatorial approaches such as using CPP with chemical platform that would activate/inhibit specific signaling cascade or vitamin C for curtailing cellular senescence [55] and also depletion of Mbd3 [56] may provide plausible insight and solutions in this regard. This will pave the way for p-iPS cell implications in basic research and biomedical applications. Moreover, the implications of CPP may go well beyond iPS cell generation with lower risk of teratoma formation [11] to their directed differentiation and gene editing [57].
Conclusion
iPS cells represent a valuable source to derive patient-specific cells for disease modeling, drug toxicity screening and cell replacement therapy applications. Introduction of exogenous genes via viral vectors, plasmids and nucleic acids in somatic cells to generate iPS cells cannot completely eliminate the possibility of genomic integration. Therefore, novel safe approaches should be explored to generate clinically safe iPS cells. In this review, we have highlighted reports that demonstrated the potency of CPP-mediated protein transduction technology, which involves delivery of transcription factors in recombinant forms into the somatic cells to generate iPS cells. Delivery of this CPP-mediated core stem cell-specific recombinant proteins can regulate gene expression, and function transiently without any genomic alteration. In addition, this protein transduction technology will help to us understand the mechanism of cell reprogramming phenomenon. This is attainable as it offers greater control over concentration and time of application. Moreover, it provides ample flexibility to experiment various combinations of reprogramming factors with small molecules using a sliding window approach to unravel their role at different stages during cell reprogramming. Importantly, this technology does not involve any genomic integration and induce an innate immune response. Henceforth, it also permits us to generate clinical grade transgene-free iPS cells for biomedical applications. Although several questions remain to be answered, and existing limitations on the use of CPP-mediated protein transduction are yet to overcome, it is clear that this evolving realistic protein transduction technology has much to offer in a cell reprogramming paradigm. This technology undoubtedly reinforces the significant role of CPPs and its immense potential in delivery of reprogramming proteins to generate transgene-free iPS cells. In conclusion, this technology is devoid of any possible risk of permanent genomic integrations and/or mutations and therefore will translate iPS cell related applications from laboratory to the clinic.
We thank all the members of Laboratory for Stem Cell Engineering and Regenerative Medicine (SCERM) for their excellent support. This work was supported by grant funded to Rajkumar P Thummer and Nibedita Lenka by North Eastern Region – Biotechnology Programme Management Cell (NER-BPMC), Department of Biotechnology, Government of India (BT/PR16655/NER/95/132/2015) and also supported by Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India (Early Career Research Award; ECR/2015/000193) funded to Rajkumar P Thummer.
- Takahashi K, Yamanaka S (2006) Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126: 663-676. Link: https://goo.gl/Th8h5T
- Worsdorfer P, Thier M, Kadari A, Edenhofer F (2013) Roadmap to cellular reprogramming–manipulating transcriptional networks with DNA, RNA, proteins and small molecules. Curr Mol Med 13: 868-878. Link: https://goo.gl/pIOc3p
- Hu K (2014) All Roads Lead to Induced Pluripotent Stem Cells: The Technologies of iPSC Generation. Stem Cells and Dev 23: 1285-1300. Link: https://goo.gl/XCX5u3
- Ben-David U, Benvenisty N (2011) The tumorigenicity of human embryonic and induced pluripotent stem cells. Nature Rev Cancer 11: 268-277. Link: https://goo.gl/CZ9t4s
- Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H et al. (2010) Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell 7: 618-630. Link: https://goo.gl/kEax8n
- Yakubov E, Rechavi G, Rozenblatt S, Givol D (2010) Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun 394: 189-193. Link: https://goo.gl/yMS9i8
- Hou P, Li Y, Zhang X, Liu C, Guan J et al. (2013) Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 341: 651-654. Link: https://goo.gl/4Qm9Xj
- Anokye-Danso F, Trivedi Chinmay M, Juhr D, Gupta M, Cui Z et al. (2011) Highly Efficient miRNA-Mediated Reprogramming of Mouse and Human Somatic Cells to Pluripotency. Cell Stem Cell 8: 376-388. Link: https://goo.gl/zbSfJO
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi Dyah L et al. (2011) Reprogramming of Mouse and Human Cells to Pluripotency Using Mature MicroRNAs. Cell Stem Cell 8: 633-638. Link: https://goo.gl/PlVYi4
- Kim D, Kim C-H, Moon J-I, Chung Y-G, Chang M-Y et al. (2009) Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell 4: 472-476. Link: https://goo.gl/H3m0xX
- Zhou H, Wu S, Joo JY, Zhu S, Han DW et al. (2009) Generation of Induced Pluripotent Stem Cells Using Recombinant Proteins. Cell Stem Cell 4: 381-384. Link: https://goo.gl/eDBYQP
- Lee J, Sayed N, Hunter A, Au Kin F, Wong Wing H et al. (2012) Activation of Innate Immunity Is Required for Efficient Nuclear Reprogramming. Cell 151: 547-558. Link: https://goo.gl/EKAYJN
- Zhang H, Ma Y, Gu J, Liao B, Li J et al. (2012) Reprogramming of somatic cells via TAT-mediated protein transduction of recombinant factors. Biomaterials 33: 5047-5055. Link: https://goo.gl/OZDFth
- Nemes C, Varga E, Polgar Z, Klincumhom N, Pirity MK et al. (2013) Generation of Mouse Induced Pluripotent Stem Cells by Protein Transduction. Tissue Engineering Part C: Methods 20: 383-392. Link: https://goo.gl/7wcj4K
- Guo Z, Peng H, Kang J, Sun D (2016) Cell-penetrating peptides: Possible transduction mechanisms and therapeutic applications (Review). Biomedical Reports. Link: https://goo.gl/K66uLj
- Madani F, Lindberg S, Langel Ü, Futaki S, Gräslund A (2011) Mechanisms of Cellular Uptake of Cell-Penetrating Peptides. Journal of Biophysics 2011: 1-10. Link: https://goo.gl/WB7qdS
- Frankel AD, Pabo CO (1988) Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55: 1189-1193. Link: https://goo.gl/Ge7bj8
- Green M, Loewenstein PM (1988) Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 55: 1179-1188. Link: https://goo.gl/5LOhjD
- Vives E, Brodin P, Lebleu B (1997) A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem 272: 16010-16017. Link: https://goo.gl/fj4Nrr
- Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF (1999) In vivo protein transduction: delivery of a biologically active protein into the mouse. Science 285: 1569-1572. Link: https://goo.gl/4RKjbm
- Ignatovich IA, Dizhe EB, Pavlotskaya AV, Akifiev BN, Burov SV et al. (2003) Complexes of plasmid DNA with basic domain 47-57 of the HIV-1 Tat protein are transferred to mammalian cells by endocytosis-mediated pathways. J Biol Chem 278: 42625-42636. Link: https://goo.gl/D323ht
- Joliot A, Pernelle C, Deagostini-Bazin H, Prochiantz A (1991) Antennapedia homeobox peptide regulates neural morphogenesis. Proceedings of the National Academy of Sciences 88: 1864-1868. Link: https://goo.gl/m82tbG
- Perez F, Joliot A, Bloch-Gallego E, Zahraoui A, Triller A et al. (1992) Antennapedia homeobox as a signal for the cellular internalization and nuclear addressing of a small exogenous peptide. J Cell Sci 102: 717-722. Link: https://goo.gl/g2XCRH
- Derossi D, Joliot AH, Chassaing G, Prochiantz A (1994) The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem 269: 10444-10450. Link: https://goo.gl/lXsSI6
- Futaki S, Ohashi W, Suzuki T, Niwa M, Tanaka S et al. (2001) Stearylated arginine-rich peptides: a new class of transfection systems. Bioconjug Chem 12: 1005-1011. Link: https://goo.gl/oWXSmG
- Matsushita M, Tomizawa K, Moriwaki A, Li S-T, Terada H et al. (2001) A high-efficiency protein transduction system demonstrating the role of PKA in long-lasting long-term potentiation. The Journal of Neuroscience 21: 6000-6007. Link: https://goo.gl/nkFT39
- Hitsuda T, Michiue H, Kitamatsu M, Fujimura A, Wang F et al. (2012) A protein transduction method using oligo-arginine (3R) for the delivery of transcription factors into cell nuclei. Biomaterials 33: 4665-4672. Link: https://goo.gl/2ypK8b
- Morris MC, Deshayes S, Heitz F, Divita G (2008) Cell-penetrating peptides: from molecular mechanisms to therapeutics. Biol Cell 100: 201-217. Link: https://goo.gl/lvk9N1
- Chaturvedi G, Simone PD, Ain R, Soares MJ, Wolfe MW (2009) Noggin maintains pluripotency of human embryonic stem cells grown on Matrigel. Cell Prolif 42: 425-433. Link: https://goo.gl/DhGevj
- Yu M, Lian S, Han H, Yu K, Li G et al. (2013) Four recombinant pluripotency transcriptional factors containing a protein transduction domain maintained the in vitro pluripotency of chicken embryonic stem cells. Science China Life Sciences 56: 40-50. Link: https://goo.gl/3OAeHy
- Peitz M, Münst B, Thummer RP, Helfen M, Edenhofer F (2014) Cell-permeant recombinant Nanog protein promotes pluripotency by inhibiting endodermal specification. Stem Cell Research 12: 680-689. Link: https://goo.gl/7khAbC
- Imsoonthornruksa S, Pruksananonda K, Parnpai R, Rungsiwiwut R, Ketudat-Cairns M (2015) Expression and Purification of Recombinant Human Basic Fibroblast Growth Factor Fusion Proteins and Their Uses in Human Stem Cell Culture. J Mol Microbiol Biotechnol 25: 372-380. Link: https://goo.gl/VU1dt9
- Onuma Y, Higuchi K, Aiki Y, Shu Y, Asada M et al. (2015) A Stable Chimeric Fibroblast Growth Factor (FGF) Can Successfully Replace Basic FGF in Human Pluripotent Stem Cell Culture. PLoS One 10: e0118931. Link: https://goo.gl/qzsD81
- Kaini RR, Shen-Gunther J, Cleland JM, Greene WA, Wang H-C (2016) Recombinant Xeno-Free Vitronectin Supports Self-Renewal and Pluripotency in Protein-Induced Pluripotent Stem Cells. Tissue Engineering Part C: Methods. Link: https://goo.gl/lH2cQ0
- Münst B, Thier MC, Winnemöller D, Helfen M, Thummer RP et al. (2016) Nanog induces suppression of senescence through downregulation of p27KIP1expression. J Cell Sci 129: 912-920. Link: https://goo.gl/iJ2BFc
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M et al. (2008) Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 26: 795-797. Link: https://goo.gl/as3SU3
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A et al. (2008) Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 26: 1269-1275. Link: https://goo.gl/GozCmY
- Lim J, Kim J, Kang J, Jo D (2014) Partial Somatic to Stem Cell Transformations Induced By Cell-Permeable Reprogramming Factors. Sci Rep 4. Link: https://goo.gl/85t9X1
- Muenthaisong S, Ujhelly O, Polgar Z, Varga E, Ivics Z et al. (2012) Generation of mouse induced pluripotent stem cells from different genetic backgrounds using Sleeping beauty transposon mediated gene transfer. Exp Cell Res 318: 2482-2489. Link: https://goo.gl/0xtGNv
- Thier M, Mnst B, Edenhofer F (2010) Exploring refined conditions for reprogramming cells by recombinant Oct4 protein. The International Journal of Developmental Biology 54: 1713-1721. Link: https://goo.gl/0PMaK9
- Thier M, Münst B, Mielke S, Edenhofer F (2012) Cellular Reprogramming Employing Recombinant Sox2 Protein. Stem Cells International 2012: 1-10. Link: https://goo.gl/bHBi5j
- Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F (2002) Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proceedings of the National Academy of Sciences 99: 4489-4494. Link: https://goo.gl/iFBsdw
- Stock K, Nolden L, Edenhofer F, Quandel T, Brüstle O (2010) Transcription factor-based modulation of neural stem cell differentiation using direct protein transduction. Cell Mol Life Sci 67: 2439-2449. Link: https://goo.gl/nPtBco
- Tang Y, Lin C-J, Tian XC (2011) Functionality and Transduction Condition Evaluation of Recombinant Klf4 for Improved Reprogramming of iPS Cells. Cellular Reprogramming (Formerly 'Cloning and Stem Cells') 13: 99-112. Link: https://goo.gl/vHlPGg
- Pan C, Lu B, Chen H, Bishop CE (2009) Reprogramming human fibroblasts using HIV-1 TAT recombinant proteins OCT4, SOX2, KLF4 and c-MYC. Mol Biol Rep 37: 2117-2124. Link: https://goo.gl/50vIRL
- Geiman DE, Ton-That H, Johnson JM, Yang VW (2000) Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein–protein interaction. Nucleic Acids Res 28: 1106-1113. Link: https://goo.gl/WAWlDY
- Caron NJ, Quenneville SP, Tremblay JP (2004) Endosome disruption enhances the functional nuclear delivery of Tat-fusion proteins. Biochem Biophys Res Commun 319: 12-20. Link: https://goo.gl/FO8YHp
- Wadia JS, Stan RV, Dowdy SF (2004) Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med 10: 310-315. Link: https://goo.gl/qmGZh3
- Bosnali M, Edenhofer F (2008) Generation of transducible versions of transcription factors Oct4 and Sox2. Biol Chem 389. Link: https://goo.gl/HhLFnO
- Yang WC, Patel KG, Lee J, Ghebremariam YT, Wong HE et al. (2009) Cell-free production of transducible transcription factors for nuclear reprogramming. Biotechnol Bioeng 104: 1047-1058. Link: https://goo.gl/TKt3yK
- Hu PF, Guan WJ, Li XC, Ma YH (2012) Construction of recombinant proteins for reprogramming of endangered Luxi cattle fibroblast cells. Mol Biol Rep 39: 7175-7182. Link: https://goo.gl/ZnVFoW
- Ryu J, Park HH, Park JH, Lee HJ, Rhee WJ et al. (2016) Soluble expression and stability enhancement of transcription factors using 30Kc19 cell-penetrating protein. Appl Microbiol Biotechnol 100: 3523-3532. Link: https://goo.gl/Hqfjxb
- Konno M, Masui S, Hamazaki TS, Okochi H (2011) Intracellular reactivation of transcription factors fused with protein transduction domain. J Biotechnol 154: 298-303. Link: https://goo.gl/wxxjOo
- Räägel H, Säälik P, Hansen M, Langel Ü, Pooga M (2009) CPP–protein constructs induce a population of non-acidic vesicles during trafficking through endo-lysosomal pathway. J Control Release 139: 108-117. Link: https://goo.gl/55HSGk
- Esteban MA, Wang T, Qin B, Yang J, Qin D et al. (2009) Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell stem cell 6: 71-79. Link: https://goo.gl/UcXGE4
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E et al. (2013) Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502: 65-70. Link: https://goo.gl/FQdfCf
- Kaitsuka T, Tomizawa K (2015) Cell-penetrating peptide as a means of directing the differentiation of induced-pluripotent stem cells. International journal of molecular sciences 16: 26667-26676. Link: goo.gl/3FPaXL .
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
